Original Research
2017
March
Volume : 5
Issue : 1
Significance of hepatitis C virus (HCV) viremia in anti-HCV antibody (IgG) positive subjects
Sukrutha Gopal Reddy, Anil Kumar Bilolikar, Prasanna Lakshmi Kakarla
Pdf Page Numbers :- 17-20
Sukrutha Gopal Reddy1*, Anil Kumar Bilolikar2, Prasanna Lakshmi Kakarla2
1Department of Molecular Biology, Krishna Institute of Medical Sciences, Minister Road, Secunderabad-500003, Telangana, India
2Department of Microbiology, Krishna Institute of Medical Sciences, Minister Road, Secunderabad-500003, Telangana, India
*Corresponding author: Dr. Sukrutha Gopal Reddy, Department of Molecular Biology, Krishna Institute of Medical Sciences, Minister Road, Secunderabad-500003, Telangana, India. Mobile: 9177111719; Email: sukruthagopalreddy@gmail.com
Received 23 September 2016; Revised 18 November 2016; Accepted 30 November 2016; Published 19 December 2016
Citation: Reddy SG, Bilolikar AK, Kakarla PL. Significance of hepatitis C virus (HCV) viremia in anti-HCV antibody (IgG) positive subjects. J Med Sci Res. 2017; 5(1):17-20. DOI: http://dx.doi.org/10.17727/JMSR.2017/5-4
Copyright: © 2017 Reddy SG, et al. Published by KIMS Foundation and Research Center. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction: Hepatitis C is one of the most common bloodborne diseases causing significant morbidity and mortality globally.
Objective: This study was designed to evaluate the correlation between 3rd generation ELISA positives for anti-HCV antibodies with real time PCR, among various categories of patients attending KIMS hospital.
Methods: This is a hospital based retrospective study at KIMS hospital, Secunderabad, India conducted between August 2013 and July 2015. From laboratory data, 115 patients, found to be anti-HCV antibody positive (done by CLIA method) were included in the study. Real-Time Reverse Transcription-PCR (RT-PCR) for hepatitis C virus (HCV) was done for these patient samples subsequently.
Results: Among a total of 115 anti-HCV antibody positive cases, 70(60.86%) were positive and 45(39.13%) were negative by HCV RNA PCR.
Conclusion: ELISA assays have many advantages in diagnostic settings including ease of automation, ease of use, relative cost effectiveness and low variability. Additional or confirmatory testing like RT-PCR is often helpful. The HCV RNA test is still regarded as a standard in the detection of active infection and for monitoring treatment.
Keywords: Anti-HCV antibody, RT-PCR, Horseradish peroxidase (HRP), Chemiluminiscent Immuno assay (CLIA).
Full Text
Hepatitis C virus (HCV) infection is a global public health problem [1]. Transmission is mostly through blood and blood products, sexual contact and from mother to newborn. High risk population includes patients receiving multiple blood transfusions, intravenous drug abusers, patients on haemodialysis, thalassemia patients, hemophiliacs and those with multiple sexual partners [2].
The hepatitis C virus is a single-stranded RNA flavivirus. It has seven major genotypes on the basis of variations in the non-structural NS-5 region of virus genome. Genotypes 3, 1, 4 & 6 are commonly reported from India. Genotypes influence progression of chronic liver disease and response to interferon therapy [3]. Hepatitis C virus infects only humans. The disease has a slowly progressive course. About 15% of HCV infected individuals recover spontaneously (viral clearance); an additional 25% have an asymptomatic illness with persistently normal aminotransferases and generally benign histological lesions. About 20% of patients with chronic hepatitis C develop cirrhosis in 10–20 years and may die of complications of cirrhosis and hepatocellular carcinoma in the absence of liver transplantation [1].
Laboratory tests that are available for the diagnosis and management of HCV include: (a) Serological tests to detect anti-HCV antibodies, (b) Molecular tests to detect and quantify HCV RNA and (c) Genotyping to detect the type of virus [4].
HCV, in its initial stages cannot be detected through current serological procedures as anti-HCV antibodies develop after 45 days of infection [5]. HCV RNA detection by molecular techniques (RT-PCR) is highly sensitive and most consistent method for the diagnosis of HCV infection in its earlier stage [6]. Determination of the anti – HCV antibodies status of patient alone is not sufficient to start antiviral therapy.
Current study is aimed to evaluate HCV viremia in anti-HCV antibody positive subjects.
Material and methods
This is a hospital based retrospective study at Krishna Institute of Medical Sciences, Secunderabad, India between August 2013 and July 2015. Samples from all departments were included in the study. From the laboratory data, 115 patients found anti – HCV antibody positive by CLIA method were taken as the study group. For these patients, HCV RNA detection was done subsequently by RT-PCR in the Molecular Biology department.
Serological assay
Anti-HCV (IgG) antibody was detected by a third generation HCV ELISA (Ortho clinical diagnostics). The ELISA HCV 3.0 uses microwells coated with a combination of recombinant HCV – encoded antigens (C22-3, C200 and NS5). HCV antibodies present in the sample binds with HCV recombinant antigens coated on the wells. After washing, Horse Radish Peroxidase (HRP) labeled antibody conjugate is added leading to a luminescent reaction. The light signals are read by VITROS system. The amount of HRP conjugate bound is directly proportional to the level of anti-HCV antibodies present.
Molecular assay
RNA was extracted from serum samples as per QIA amp viral RNA purification protocol (QIAGEN GmbH) using stringent precautions to avoid contamination. Viral load detection was done by reverse transcription polymerase chain reaction (RT-PCR) in Rotor-Gene Q. The specific amplification of 240bp region of the HCV genome is done & detected by fluorescent dyes linked to specific probes.
Quantitative standards and negative control are included in the PCR assay. Monitoring the fluorescence intensities during the PCR run allows the quantitation of accumulated HCV RNA. Detection limit of the kit is 65-106 IU/mL.
Results
Of the total 115 anti-HCV antibody positive samples by CLIA, 91 (79.17%) were males and 24 (20.8%) were females. Majority of the patients are males in the middle age group 41-60 years (Figure 1).
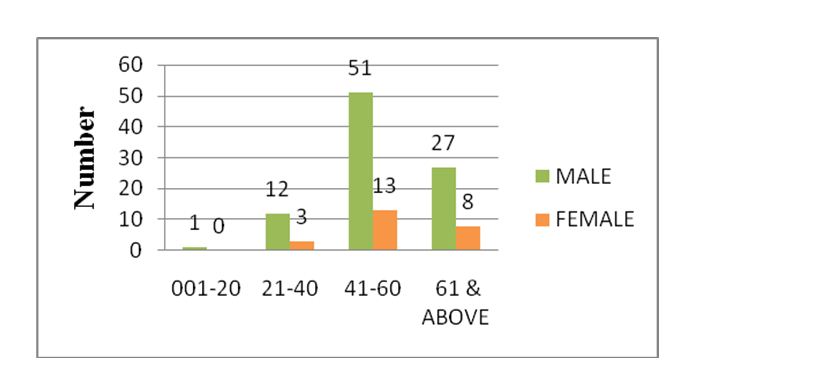
Figure 1: Age & sex distribution of anti-HCV antibody positive cases.
All the positive samples were subjected to quantitative real time PCR and the results are depicted in Figure 2.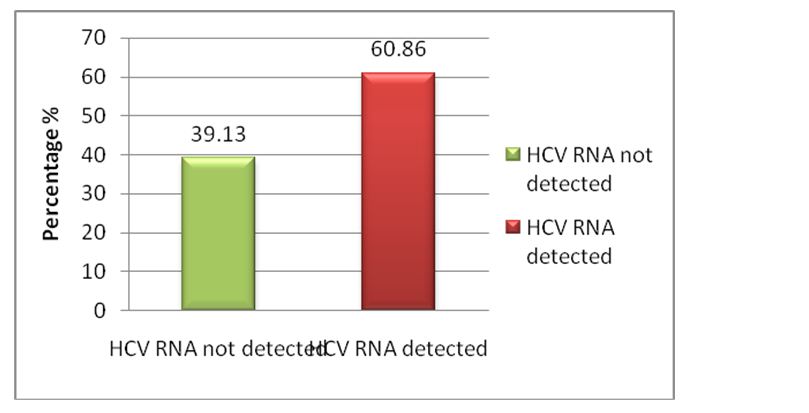
Figure 2: Positivity and negativity by RT-PCR.
Out of 115 anti-HCV antibody positive cases, 70 (60.86%) were positive and 45 (39.13%) were negative for HCV RNA by RT-PCR. Among 70 HCV RNA positive cases, 61 (67.03%) were males and 09(37.5%) were females.
Discussion
Hepatitis C is an emerging infection in India with long term implications. It is a pathogen that is already responsible for a significant proportion of liver disease in various regions of India [7]. The natural history of hepatitis C infection is depicted in Figure 3.
Figure 3: Natural history of hepatitis C infection [8].
Abbreviations: HCC – Hepatocellular compensation; decomp - Decompensation
In early phase of acute HCV infection, there is a window period. During this period the antibodies are not detectable by ELISA. Hence the ELISA test will be falsely negative despite the viremia [2]. False negative tests are also frequent in those patients who are immunodeficient due to chemotherapy, steroid therapy, HIV coinfection, immunosuppressive therapy for organ transplantation, chronic renal failure who are on haemodialysis and neonates being assessed for vertical transmission [2]. In such cases, HCV RNA detection is the method of choice.
Moreover, there is role of RT-PCR even in anti-HCV antibody positive cases to confirm ongoing infection, as immunoassays are not confirmatory. The HCV RNA test is regarded as standard in the detection of active infection and also for monitoring treatment. Baseline HCV RNA load is an independent predictor of treatment success during interferon based therapy. Genotyping for further identification of distinct strains of different geographic areas is recommended. Recommended testing sequence for identifying current hepatitis C virus infection as given by CDC [9] is shown in Figure 4.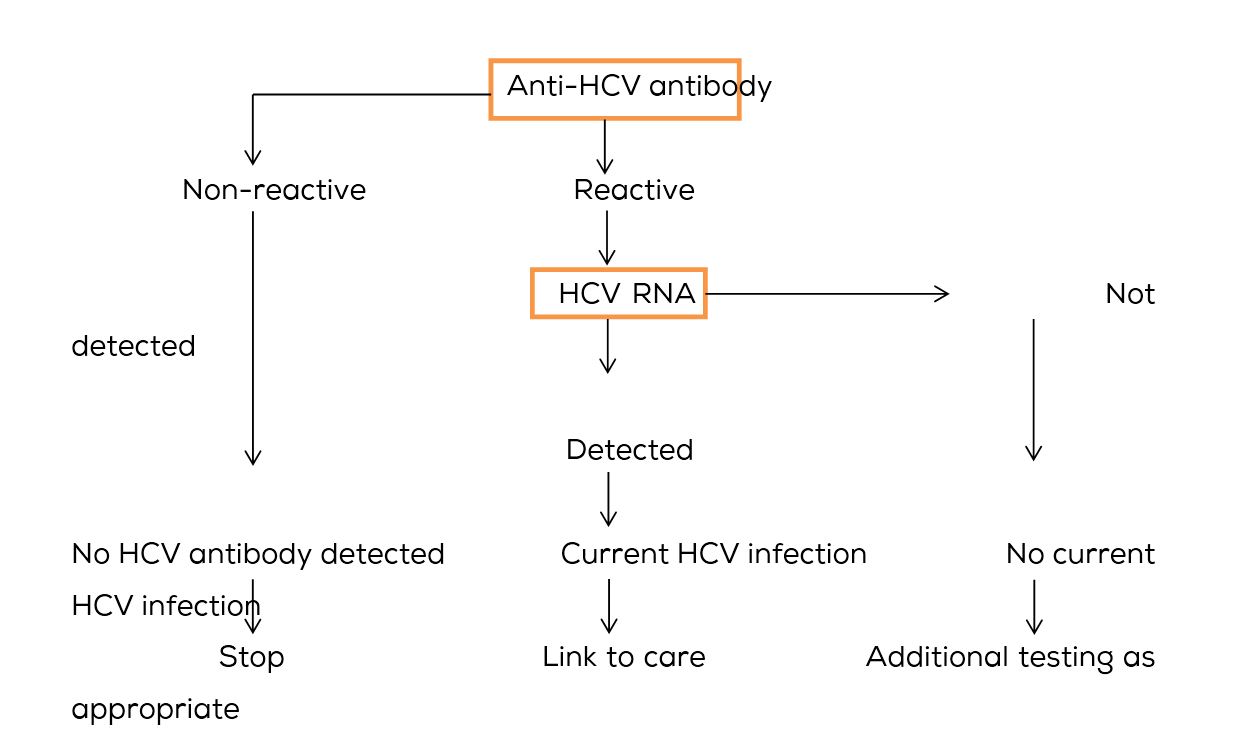
Figure 4: Recommended testing sequence for identifying current hepatitis C virus infection as given by CDC [9].
In this study, 60.86% cases had active hepatitis C virus infection indicating chronicity. A 77% concordance between serological and PCR results was found in their study titled “Antibody testing and RT-PCR results in hepatitis C virus infection: HCV-RNA detection in PBMC of plasma viremia-negative HCV-seropositive persons”, by Caudai C et al. [10]. However, 18% of samples were positive for antibody and negative for RNA.
Results comparable to our study were observed in a similar study done by Abida A et al. [11] in our neighboring country Pakistan, where 37.5% cases were negative and 62.5% were positive by RT-PCR among anti-HCV antibody positive cases. Another study done by Anwar Ali et al. [12] showed 16.92% negativity by RT-PCR which is far less when compared to our study.
Datta S et al. recommend regular routine screening of HCV by PCR in ESRD patients to help in early diagnosis of HCV infection [13]. In their study population (ESRD patients), PCR positivity (67.4%) was quite high when compare to HCV serology positivity (36.5%). ESRD patients are a known high-risk group for acquiring HCV infection. Our study included all categories of patients and hence the difference in results can be explained.
Conclusion
Enzyme immunoassays are done for routine screening of HCV infection but they do not distinguish among acute, chronic or resolved infection. HCV RNA in blood is an early predictor of virus induced liver damage in anti-HCV antibody positive patients. We recommend HCV RNA quantitative estimation for all anti-HCV antibody positive patients to confirm active infection as high proportion of these patients may progress to chronic liver disease. HCV PCR is also recommended for anti-HCV antibody negative patients who are at high risk for acquiring the infection. Further studies on hepatitis C virus genotype detection facilitating improved clinical outcome and epidemiology are required.
Acknowledgements
Krishna Institute of Medical Sciences (KIMS), Secunderabad-500003, Telangana, India.
Conflicts of interest
Authors declare no conflicts of interest.
References
[1] Kurtulus B, Gulcin B, Nese K, Zeynep A, Selcuk K, et al. Correlation between architect hepatitis C virus (HCV) core antigen and HCV Ribonucleic acid levels in Anti-HCV reactive patients in Turkey. Glob J Med Microb Rev. 2013; 1(1):1-5.
[2] Swellam M, Mahmoud MS, Ali AA. Diagnosis of hepatitis C virus infection by enzyme-linked immunosorbent assay and reverse tanscriptase-nested polymerase chain reaction: A comparative evaluation. IUBMB Life. 2011; 63(6):430-434.
[3] Naghmi A, Tahira Z, Khalid H, Lubna N. Seroprevalence anti HCV antibodies, HCV-RNA and its genotype among patients of hemophilia, at hemophilia treatment center Pakistan Institute of Medical Sciences, Islamabad. International Journal of pathology. 2009; 7(2):84-87.
[4] Ashis M. Hepatitis C in India. J Biosci. 2008; 33(4):465-473.
[5] Ayesh BM, Zourob SS, Abu-Jadallah SY, Shemer Y. Most common genotypes and risk factors for HCV in Gaza strip: a cross sectional study. Virol J. 2009; 6:105-112.
[6] World Health Organization. Hepatitis C. WHO, Geneva. 2002.
[7] Tashkandya MA, Khodari YA, Ibrahaim AM, Dhafar KO, Gazzaz ZJ, et al. Evaluation of the available anti-HCV antibody detection tests and RT-PCR assay in the diagnosis of Hepatitis C Virus infection. Saudi J Kidney Dis Transpl. 2007; 18(4):523-531.
[8] Poynard T, Ratziu V, Benhamou Y, Opolon P, Cacoub P, et al. Natural history of HCV infection. Baillieres Best Pract Res Clin Gastroenterol. 2000;14:211-228.
[9] Centers for Disease Control and Prevention (CDC). Testing for HCV infection: An update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013; 62(18):362-365.
[10] Caudai C, Padula MG, Bastianoni I, Almi P. Antibody testing and RT-PCR results in hepatitis C virus (HCV) infection: HCV-RNA detection in PBMC of plasma viremia-negative HCV-seropositive persons. Infection. 1998; 26(3):151-154.
[11] Abida A, Habib Ullah Khan, Farzana, Mehwish SA, Tayyab R, et al. Seropositivity and active HCV infection in patients from Peshawar division of Khyber Pakhtunkhwa province. Pakistan J. 2001; 45(6):1579-1584.
[12] Ali A, Lal A. False positivity of serological tests for Hepatitis C virus. J Ayub Med Coll Abottabad. 2010; 22(2):43-45.
[13] Datta S, Goel N, Wattal C. Utility of routine real time quantitative PCR monitoring of HCV infection in haemodialysis patients. Indian J Med Microbiol. 2015; 33(5):106-111.